一 : in case 和 in case of 的区别
in case 和 in case of 的区别
莘莘你怎么干这么诡异的事啊
把BAIDU上的答案贴给你
in case万一.是连词,引导条件状语从句.也就是说in case后面是一个完整的句子 Take a hat with you in case the sun is very hot.倘若太阳很利害,你就把帽子戴上.in case of的of是介词,介词后面只能带名词性质的词,比如名词、代词等.意思和in case差不多,万一的意思 In case of rain they can't go.万一下雨,他们就不能去了.in the case of,就...来说,关于.意思跟上面两个不一样了.一般表示转而提及另一件事情.比如 In the case of woman,they have more difficulty in their job.就女性来说,她们在工作中会遇到更多的困难
1.We have an auxiliary generator in case of power cuts.
我们有一台万一断电时使用的备用发电机.
2.In case you need something,please don't hesitate to let me know.
如果你需要什么东西,请不客气地对我说.
3.It may rain you'd better take an umbrella (just) in case (it does).
可能下雨--你最好带把伞,以防万一(下起来).
4.In case that he leaves,please inform me.
如果他离开,请通知我.
5.In case of rain,they can't go.
万一下雨,他们就不能去了.
6.In case (=If) I forget,please remind me.
万一我忘记,请提醒我.
7.Write the telephone number down in case you forget.
把电话号码写下来以免忘了.
8.The doctor asked us to call him during the night except in case of necessity.
医生吩咐我们,除非必须,否则不要在夜里叫他.
简单来说,in case+从句 in case of+名词性质的词
二 : in case/in the case/in case of的用法及区别
incase万一。是连词,引导条件状语从句。也就是说incase后面是一个完整的句子 Take a hat with you in case the sun isvery hot. 倘若太阳很利害,你就把帽子戴上。 in caseof的of是介词,介词后面只能带名词性质的词,比如名词、代词等。意思和incase差不多,万一的意思 In case of rain they can't go.万一下雨,他们就不能去了。 in the case of,就...来说,关于。意思跟上面两个不一样了。一般表示转而提及另一件事情。比如 Inthe case of woman,they have more difficulty in theirjob。就女性来说,她们在工作中会遇到更多的困难一、case作名词的用法:三 : Characterization of T-DNA insertion patterns in the g
Curr Genet (2007) 51:233–243 DOI 10.1007/s00294-007-0122-5
RESEARCH ARTICLE
Characterization of T-DNA insertion patterns in the genome of rice blast fungus Magnaporthe oryzae
Guihua Li · Zhuangzhi Zhou · Guifu Liu · Fucong Zheng · Chaozu He
Received: 11 December 2006 / Revised: 27 January 2007 / Accepted: 29 January 2007 / Published online: 16 February 2007? Springer-Verlag 2007
AbstractAgrobacterium tumefaciens-mediated trans-formation (ATMT) has been proven to be a powerfulstrategy for gene disruption in plants and fungi. Pat-terns associated with transferred DNA (T-DNA)integration in plants and yeast have been studiedcomprehensively, whereas no detailed analysis ofT-DNA integration has been reported yet in Wlamen-tous fungi. Here, we reported the T-DNA insertionpatterns in the genome of Wlamentous fungus Magna-porthe oryzae. Using ATMT, a T-DNA tagged popula-tion consisting of 6,179 transformants of M. oryzae wasconstructed. With thermal asymmetric interlaced-PCR(TAIL-PCR), 623 right border (RB) Xankingsequences and 124 left border (LB) Xanking sequenceswere generated. Analysis of these Xanking sequencesindicated a signiWcant integration bias toward non-cod-ing sequences, suggesting distribution of T-DNAswas not random. Comparing to T-DNA RB, LB wasnicked inaccurately and truncated frequently during
integration. Chromosomal rearrangements, such asdeletion, inversion, and translocation, were associatedwith T-DNA integration in some transformants. Ourdata suggest that, comparing with plant cells, T-DNAintegrates into this Wlamentous fungus with more pre-cise and simpler patterns. Some phenotypic mutantswere observed in our T-DNA tagged population, andthese transformants will be very useful for functionalgenomics research of M. oryzae.
KeywordsAgrobacterium tumefaciens-mediated transformation · Magnaporthe oryzae · T-DNA · Chromosomal rearrangement
Introduction
Magnaporthe oryzae (formerly Magnaporthe grisea),the causative agent of rice blast, is the most destructivepathogen of rice worldwide (Zeigler etal. 1994) andthe principal model organism for elucidating themolecular basis of fungus-plant interactions (Valent1990). M. oryzae is a haploid ascomycete fungusandcontains seven chromosomes in each cell. Thedraft genome sequence of M. oryzae strain 70-15 hasbeen released (Dean etal. 2005). Functional and com-parative genomic analysis of this important phytopath-ogenic fungus, although still in the early stage, hasimproved our understanding of the molecular mecha-nisms involved in host-pathogen interactions (Xu etal.2006). Large-scale expressed sequence tags (EST),whole-genome microarrays and mutant collectionshave been developed (Soanes etal. 2002; Xu etal.2006). Among available approaches for Wnding novelgenes that encode determinants of pathogenicity,
Communicated by K. Borkovich.
Guihua Li and Zhuangzhi Zhou contributed equally to the work.Electronic supplementary materialThe online version of this article (doi:10.1007/s00294-007-0122-5) contains supplementary material, which is available to authorized users.
G. Li · Z. Zhou · G. Liu · F. Zheng · C. He (&)State Key Laboratory of Plant Genomics,
Institute of Microbiology, Chinese Academy of Sciences, Beijing 100080, People’s Republic of Chinae-mail: hecz@im.ac.cn
G. Li · Z. Zhou
Graduate School of the Chinese Academy of Sciences, Beijing 100039, People’s Republic of China
123
234identiWcation of pathogenicity-deWcient mutantsclearly has the greatest potential. Agrobacteriumtumefaciens-mediated transformation (ATMT) hasbeen proved to be an eVective way for this aim inplants. Transferred DNA (T-DNA) transformation ofM. oryzae was demonstrated successfully recently (Rhoetal. 2001).
Agrobacterium tumefaciens is able to transfer itsT-DNA into diverse groups of organisms, includingplants, other prokaryotes, yeast, Wlamentous fungi,even human cells, which represents a unique case ofnaturally occurring trans-kingdom DNA transfer (Lac-roix etal. 2006). Whereas extensive work has revealedthe translocation process, little is known about the bio-logical mechanisms governing the targeting of the T-DNA to its site of integration and following insertionwithin the host chromatin. Comprehensive character-izations associated with T-DNA insertion into plantshave been reported recently, especially in Arabidopsis,tobacco, and rice (TzWra etal. 2004).
Transferred DNA integrates into plant cells mostlywith a non-homologous recombination mode (Gheysenetal. 1991; Mayerhofer etal. 1991), and a signiWcant biasof integration events in 5?-up-stream (5? untranslatedregion and promoter) regions of genes has been dis-closed (Alonso etal. 2003; Chen etal. 2003; Forsbachetal. 2003; Sha etal. 2004). At regions surroundingcentromeres of Arabidopsis, T-DNA integration eventsare observed rarely (Alonso etal. 2003).
In Agrobacterium, the 25bp T-DNA border repeatsequence is predominantly nicked at the T-DNA lowerstrand, between the third and fourth nucleotide (Sta-chel etal. 1986; Wang etal. 1987). T-DNA integratesinto plant cells with a more canonical right border(RB) than left border (LB) (Brunaud etal. 2002; Fors-bach etal. 2003; Kim etal. 2003), probably due to theprotection of VirD2 protein which has been attachedto RB for most of the time during the process of T-DNA transfer (Tinland etal. 1995; Brunaud etal.2002).
Transferred DNA insertions in Arabidopsis (Ghey-sen etal. 1991; Mayerhofer etal. 1991; Forsbach etal.2003) and tobacco (Ohba etal. 1995) are normallyassociated with small (<100bp) target site deletions.Chromosomal rearrangements such as inversion andtranslocation, associated with T-DNA insertions, arealso observed in transgenic Arabidopsis (Castle etal.1993; Nacry etal. 1998; Forsbach etal. 2003) with lowfrequency.
At the junction between T-DNA and the plantgenomic DNA, Wller sequence (Forsbach etal. 2003;Windels etal. 2003) and microhomology are frequentlyobserved, whereas the frequency of microhomology is
123
Curr Genet (2007) 51:233–243
found higher at the junctions involving LB than thoseof RB (Forsbach etal. 2003; Kim etal. 2003).
Long T-DNA (T-DNA and non-T-DNA) transferis a very common phenomenon occurred during theprocess of ATMT of plants (Virts and Gelvin 1985;Wenck etal. 1997; Yin and Wang 2000; Sha etal.2004). Insertions with a size of greater-than-unit-length of the binary plasmid have been detected intransgenic Arabidopsis (Wenck etal. 1997) and rice(Yin and Wang 2000). Based on analysis of co-trans-formation with two diVerent T-DNAs containing dis-tinct sequences adjacent to T-DNA borders, it isconcluded that T-DNAs frequently integrate intosame positions of plant genomes with inverted- ordirect-repeat modes (De Neve etal. 1997; De Bucketal. 1999).
Recent studies indicate that T-DNA integration intodouble-stranded break (DSB) with a double-strandedform might be a default mode of T-DNA insertion inhost genomes (K?hler etal. 1989; Chilton and Que2003; TzWra etal. 2003; TzWra etal. 2004).
Agrobacterium tumefaciens-mediated transforma-tion has been established in many Wlamentous fungiincluding M. oryzae (Michielse etal. 2005). However,patterns of T-DNA insertion in these fungal genomesare not clear. In this paper, the T-DNA integrationpatterns into the genome of M. oryzae are explored.We constructed a T-DNA tagged population ofM.oryzae containing 6,179 transformants, from which623 RB Xanking sequences and 124 LB Xankingsequences were rescued and analyzed. Our resultsshowed that distribution of T-DNAs among sevenchromosomes of M. oryzae was not random, andT-DNA integration could induce chromosomal rear-rangements such as deletion, inversion, and transloca-tion. Our data suggest that T-DNA integrates into thegenome of M. oryzae with a more precise and simplermode than that into plant cells, which provides advan-tages for study of functional genomics of the Wlamen-tous fungus.
Materials and methods
Fungal strains and culture conditions
Two strains of M. oryzae were used in this study, aChinese Weld strain Y34 was used in ATMT and alaboratory strain 70-15 was used as a reference strain.The fungus was cultured on oatmeal agar (OMA) at28°C. For conidiation, the 5–7days plate culture wasscratched, followed by additional 3–4days cultivationunder continuous Xuorescent light.
Curr Genet (2007) 51:233–243Agrobacterium-mediated transformation of Magnaporthe oryzae
Agrobacterium tumeWciens strain Agl-1, carrying thebinary vector pBHt2 with hpt (hygromycin phospho-transferase) gene under the Aspergillus nidulans trpCpromoter (Mullins etal. 2001), was used to transformstrain Y34 with the protocol as previously described(Rho etal. 2001). Hygromycin B (200??g/ml) was usedfor selection of transformants.
Mitotic stability of T-DNA inserted in the Magnaporthe oryzae genome
Five randomly selected transformants were cultured onOMA plates without hygromycin B for 4–5days.Mycelia at the edge of the culture was picked up withtoothpick and inoculated on fresh OMA plates forgrowth of another 4–5days. After repeating this proce-dure for 16 times, germinating monoconidium fromeach transformant was transferred to OMA plate con-taining hygromycin B (200??g/ml) and their growth wasobserved. DNAs of each strain were extracted forSouthern blot analysis with the probe ampliWed withprimers HPTa and HPTb (Supplementary Table S1),which were speciWc to hpt gene and with an 858bpampliWcation product.DNA isolation and analysis
Following growth on OMA plates for 4–7days, myceliaof each strain were collected and cultured in V-8 juicemedia with hygromycin B (75??g/ml) and streptomycin(100??g/ml) for additional 2–4days. Then, mycelia wereharvested by vacuum-inWltration and washed by sterilewater. Mycelia were ground to Wne powders in liquidnitrogen. Genomic DNA was isolated as previouslydescribed (Talbot etal. 1993).
For Southern blot analysis, about 1??g of genomicDNA was digested with restriction endonucleases, sep-arated in 1% agarose gels, blotted to Hybond-N+ mem-branes and hybridized with [32P]dCTP-labeled probe.Hybridization and washes were performed using stan-dard procedures (Sambrook etal. 1989).Isolation of T-DNA Xanking sequences
Thermal asymmetric interlaced-PCR (TAIL-PCR)(Liu and Whittier 1995; Mullins etal. 2001) wasemployed to obtain genomic DNA sequences Xankinginserted T-DNAs from transformants selected ran-domly. Primers designed for TAIL-PCR were list inSupplementary Table S1. The tertiary TAIL-PCR
235
products were cloned into pGEM-T Easy vector (Pro-mega, Madison, WI, USA) and sequenced.Sequence processing
Sequences of TAIL-PCR products were searched fornucleotide similarities by the BLASTN algorithmagainst databases as follows: M. oryzae genomedatabase (http://www.broad.mit.edu/annotation/genome/magnaporthe_grisea) and NCBI (http://www.ncbi.nlm.nih.gov/BLAST). Positions of the T-DNA insertionsites were mapped in silico onto M. oryzae chromo-somes based on the physical and genetic maps availableat the M. oryzae genome database.
Distribution of T-DNA insertion sites among sevenchromosomes was analyzed according to length of eachlinkage group released from the latest Assembly Struc-ture of the M. oryzae genome database. Distribution ofT-DNA insertion sites within particular genetic ele-ments was analyzed according to data derived fromGenome Statistics of the M. oryzae genome database.Searches for 1kb up-stream or 200bp down-stream ofannotated genes of M. oryzae were preformed for dis-tribution of T-DNA insertion sites in the 5?- or 3?-regu-latory regions.
Nicking sites of T-DNA integrations were deter-mined by comparison of each end sequence of the inte-grated T-DNA, which was revealed by Xankingsequence analysis, with the end sequence of canonicalone.
Plasmid rescue
About 1??g of genomic DNA of each tested transfor-mant was digested with restriction endonuclease PstI,which has only one digestion site in plasmid pBHt2 atits multiple cloning site. The digested DNA was etha-nol precipitated, dissolved in sterile water and ligatedusing T4 DNA ligase at 16°C overnight. The product ofligation was precipitated, redissolved in 20??l sterilewater and electroporated into Escherichia coli DH10Bcells. Transformants were cultured on LB plates withkanamycin (50??g/ml) for plasmid selection.Analysis of chromosomal rearrangement
Each putative transformant, involving rearrangementprobably revealed by TAIL-PCR, was further analyzedby PCR using four sets of primer pairs designedaccording to the surrounding sequences of junctionsites (primers LF1-3 and LR1-3 for LB junction sites,RF1-3 and RR1-3 for RB junction sites, Supplemen-tary Table S1). PCR products containing host–host
123
236Fig.1

Southern blot analysis of transformants of Magnaportheoryzae. Genomic DNAs of diVerent transformants and wild typestrain Y34 were digested with PstI and hybridized with a fragment
junction sites were sequenced to verify the rearrange-ments. In order to further conWrm chromosomal inver-sion occurred in a transformant, Southern blot analysiswith six probes derived from rearranged chromosomalregions was carried out.
Results
Determination of T-DNA copy number and mitotic stability of transformants of Magnaporthe oryzaeAn ATMT system of M. oryzae with transformationeYciency about 300 transformants per 1£106 conidia,was established, from which 6,179 independent trans-formants were obtained. To determine T-DNA inte-gration and copy number, 135 transformants wereselected randomly. PCR analysis with primers HPTaand HPTb indicated that all tested transformants con-tained Hpt gene (data not shown). Genomic DNAs ofthese transformants were analyzed by Southern blotprobed with a fragment of Hpt gene (Fig.1). The resultindicated that 72.6% (98/135) of the transformantscontained one copy of T-DNA, 19.2% (26/135) con-tained two copies, 8.2% (11/135) contained three ormore copies. The average copy number of T-DNAintegrated into these transformants was 1.36. Five ran-domly selected transformants were assayed for theirmitotic stability of inserted T-DNA. These trasnfor-mants remained their hygromycin B resistance and T-DNA copy number in subsequent sub-culture for 16times in the absence of hygromycin B, indicating thatT-DNA was stable in the genome of M. oryzae.Analysis of T-DNA Xanking sequences
Thermal asymmetric interlaced-PCR was carried outto recover Xanking sequences of inserted T-DNAs.Excluding the non-speciWc or similar, we obtained 623RB Xanking sequences from 900 transformants and 124LB Xanking sequences from 240 transformants, respec-
123
Curr Genet (2007) 51:233–243
of hpt gene. Lane 1, 1kb DNA marker (New England Biolabs);lanes 2–31, diVerent transformants; lane 32, wild type strain Y34
tively. From them, genomic sequences adjacent to bothborders of a same T-DNA were recovered from 75transformants. Of these 623 RB Xanking sequences,445 corresponded to single copy genomic regions; 16matched to multiple genomic regions, suggesting thatthe corresponding T-DNAs were integrated into repet-itive sequence regions; 150 Xanking sequences con-tained sequences derived from binary vector backboneor another T-DNA (of which M. oryzae sequencescould be found in three Xanking sequences); and 14Xanking sequences were not matched to any sequencesof M. oryzae or binary vector pBHt2, which mightlocate in un-sequenced regions.
Distribution of T-DNA insertion sites among seven chromosomes
The 445 solely matched Xanking sequences wereanalyzed for distribution of T-DNA insertion sites.Of which, 432 Xanking sequences were mapped onseven chromosomes, respectively, while 13 Xankingsequences located on unlinked surpercontigs (Fig. 2,Supplementary Table S2). According to length of eachchromosome available at the M. oryzae genome data-base, expected frequencies of T-DNA integrations intoseven chromosomes from I to VII based on randominsertion are 21.0, 16.0, 15.8, 10.6, 13.9, 11.7, and11.0%, respectively. Expected T-DNA numbers dis-tributed among seven chromosomes were calculatedfor the mapped 432 Xanking sequences (Table1). Theresults revealed that the distribution of T-DNA inser-tion sites among seven chromosomes was not com-pletely random. Although no signiWcant bias (atP=0.05 level) of T-DNA integration events into chro-mosomes II, IV, V, VI, and VII were observed, T-DNA insertions toward chromosome I appeared morefrequent than expected. However, for chromosome III,the frequency of T-DNA integration was signiWcantlylower than expected. Fewer insertion events of T-DNAwere found at some inner regions where centromeresmight locate (Fig.2).
Curr Genet (2007) 51:233–243Fig.2
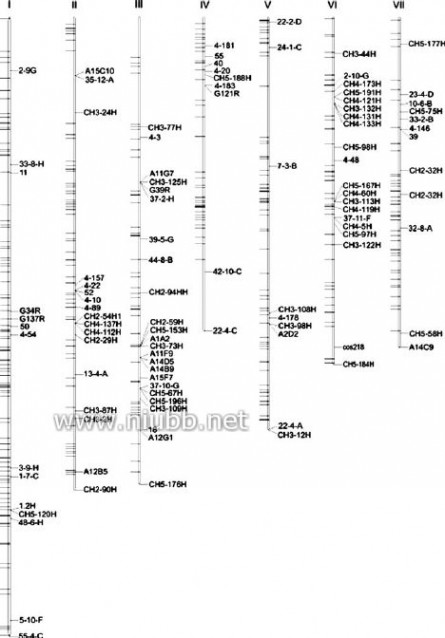
Scheme of distribution of T-DNAs in the genome of Magnaporthe oryzae.
Chromosomal positions of 432 T-DNA RB Xanking sequences were determined according to the physical and genetic maps available at the Magnaporthe oryzae genome database. The loci of corresponding T-DNA
positions were shown on left sides of each chromosome. Genetic markers of
Magnaporthe oryzae were shown on right
237
Table1Statistical analysis of the distribution of T-DNAs among seven chromosomes
ChromosomesI
Expected T-DNA numberObserved T-DNA number
90.75109*
II69.0769
III68.0948**
IV45.6740
V60.0867
VI50.5853
VII47.7646
432432Total
Exact integration sites of these T-DNAs can be found in Supplementary Table S2
??2 values were calculated for 432 T-DNAs, which map positions on chromosomes were determined unambiguously*, ** Indicate signiWcance for ??2 test at P=0.05 and 0.01 levels, respectively
Distribution of T-DNA insertion sites within particular genetic elements
Genetic elements were deduced from gene predictionsof the latest release of the M. oryzae genome annota-tion (Version 5.0). In the genome, a gene was found, onaverage, every 3,241.5bp including 1,549bp of genicsequences (1,319bp in exons and 230bp in introns).Assuming completely random T-DNA integration wewould expect 47.8% of the insertions in Coding
123
238
Fig.3

Nicking positions of the integrated T-DNAs.
Histogram of the percentage of nicking site was classiWed based on the nicking fre-quency calculated from 124 LB or 558 RB Xanking se-quences, respectively. T-DNA border repeats were capital-ized. Arrows indicate the posi-tions of T-DNA nicking in Agrobacterium
Table2Distribution of T-DNA insertion sites within particulargenetic elementsPosition of insertionExpected number Observed number of T-DNAof T-DNANon-genic sequences232.35364**Up-stream 1,000bp137.28186**Down-stream 200bp27.4620Intergenic sequences67.61159**Genic sequences212.6580**Exon181.0654**Intron
31.59
26Total sequences analyzed
445
** Indicates signiWcance at P=0.01 level for ??2 tests
sequences (40.7% in exons and 7.1% in introns), 30.8%in the region 1kb up-stream of a start codon, and 6.2%in the region 200bp down-stream of a stop codon. Theremainder (15.2%) should insert in intergenic regions.??2 tests (P=0.01) indicated that T-DNA insertions ingenic sequences were found less frequently thanexpected, mostly due to the low frequency of insertionin exon. However, a signiWcant insertion bias wasobserved in favor of non-genic sequences, especiallyup-stream 1kb regions and intergenic regions. No sig-niWcant integration biases were observed in introns ordown-stream 200bp regions of annotated genes(Table2).
Integration accuracy of T-DNA borders
Five hundred and Wfty-eight RB Xanking sequences (of623, the remainder 65 Xanking sequences belong tolong T-DNA integration) and 124 LB Xankingsequences were analyzed for integration accuracy of T-DNA borders (Fig.3). The results showed that theaccuracy of integration was very high at RB. In 92.1%(514 of 558 Xanking sequences) of the cases, T-DNAsintegrated with canonical RBs (i.e., nicking positionsappear between the third and fourth nucleotide of the25bp border repeat sequence), while deletions withsmall fragment (1–80bp) were only found in 7.9% (44of 558 Xanking sequences) of the cases. Compared with
123
Curr Genet (2007) 51:233–243
RB, however, the accuracy of LB integration was low.Only 32.3% (41 of 124) were canonical insertions. T-DNA integrations with LBs truncated with 1–22bpwere observed frequently (59.7%, 73 of 124). Largedeletions (30–150bp) occurred in lower frequency(8.1%, 10 of 124).
Microhomologies and Wller sequences observed at the junctions between the host genome and T-DNA borders
The 124 LB and the 462 RB Xanking sequences, whichcontained sequences of M. oryzae, were analyzed forpatterns of junctions between T-DNA borders andgenomic target sites (Table3). Microhomologies with2–7bp in size were found frequently (72.6%, 90 of 124)in junctions between LB and genomic sequences. How-ever, in junctions involving RB, microhomologies werefound less frequently (21.0%, 97 of 462) and mostly insmaller size (1bp). Meanwhile, Wller sequences rangingfrom 1 to 230bp in size were observed from 56 of 462(12.1%) junctions of RB. Sequence analysis showedthat, some longer Wller sequences were originated fromthe T-DNA region, backbone of binary vector or thehost genome, respectively. It was diYcult to determinethe originations of the shorter Wller sequences. At thejunctions of LB, Wller sequences occurred rarely (1.6%,2 of 124).
Sequence analysis of T-DNA insertion sites and chromosomal rearrangements associated with T-DNA integration
From RB and LB Xanking sequences mentionedabove, genomic sequences adjacent to both borders of asame T-DNA were obtained from 75 transformants.These Xanking sequences were compared withsequences corresponding to pre-insertion sites (Table4).The results indicated that, deletions with 1–80bp in size(15bp in average) were observed frequently (60 of 75)in genomic target sites due to T-DNA integration. Thelargest deletion with 505bp in size was found in one
Curr Genet (2007) 51:233–243
Table3Sequence analysis of junctions between T-DNA borders and genomic target sites
239
RBSize (bp)
Identical nucleotides between target site and border1–512–5Filler nucleotides between target site and border1–2301–3030–230
Exact joining between target site and borderTotal sequences analyzed
Table4Sequences analysis of T-DNA insertion sites
Number of T-DNA insertions
Duplication of target site1–8bp
5 (6.7%)Exact integration into target site6 (8.0%)Deletion of target site61 (81.3%)1–20bp46 (61.3%)21–80bp14 (18.7%)>80bp1 (1.3%)Inversions2 (2.7%)Translocations
1 (1.3%)Number of sequences analyzed
75(100%)
transformant. Short duplications (1–8bp) of the targetsequences were also observed in Wve T-DNA insertionevents. Exact integration without deletion or duplica-tion was only found in six transformants. These resultssuggest that small fragment deletion of the target site isthe most frequent event during T-DNA integration intothe genome of M. oryzae.
Three single-copy T-DNA transformants (6001,4825, and 438), which had RB and LB Xankingsequences localized on diVerent chromosomes, or onthe same chromosome but far away from each other,were identiWed, which suggested that chromosomalrearrangement might occur. PCR was applied to fur-ther analysis, with primers (Supplementary Table S1)speciWc to corresponding target sites of each transfor-mant. AmpliWed products were only obtained fromgenomic DNA templates of wild type strains usingprimer pairs LF+LR (LF1+LR1, LF2+LR2, andLF3+LR3 for transformant 6001, 4825, and 438,respectively) or RF+RR (RF1+RR1, RF2+RR2,and RF3+RR3 for transformant 6001, 4825 and 438,respectively) (Fig.4a–c). However, when primer pairsLF+RR (LF1+RR1, LF2+RR2, and LF3+RR3,respectively) or RF+LR (RF1+LR1, RF2+LR2,and RF3+LR3, respectively) which had annealingsites on diVerent fragments involved in rearrangement,
LBNumber of junctionsSize (bp)Number of junctions971–79070112272–778561–2
2332330932462
124
were used, PCR products with predicted sizes wereampliWed from all three transformants, while no prod-uct was obtained from wild type strains (Fig.4a–c).These results demonstrated that chromosomal rear-rangements induced by T-DNA integration occurred inthese transformants. The rearrangements were furtherconWrmed by sequencing the host–host rejoining sitesfor each transformant. Taking these results together,translocation occurred in transformant 438, with largefragments translocated between chromosomes I andIV (Fig.4d), while inversions occurred in transformant6001 and 4825, involving fragments with 64 and 218kbin size, respectively (Fig.4e).
Southern blot analysis of transformant 6001 usingsix probes derived from representative locations ofchromosomal fragments involved in arrangement,further conWrmed the inversion event (Fig.5).Fig.5IIa, e, f indicated that no duplication presentedin the T-DNA Xanking regions since only one bandwas observed for 6001 consistently. Fig.5IIc, dshowed that genomic DNAs between the two pre-dicted T-DNA insertion sites (Fig.5I) remainedintact. Fig.5IIb indicated that there was anotherrejoining site (host–host) existed somewhere in thegenome of 6001. Therefore, together with evidencesof PCR analysis and host–host junction sequencingmentioned above, chromosomal inversion event wasconWrmed further in transformant 6001.
Analysis of sequences of rearrangemental junctionregions (including host-LB, RB-host, and host–host)revealed that rejoining patterns similar to onesdescribed above for general T-DNA integration(Tables3, 4) were observed. Small fragment deletionsand microhomologies were found frequently. Dele-tions of host genomic sequences with 30, 12, and 22bpin size were observed in transformant 6001, 4825, and438, respectively. Microhomologies with 1–3bp werefound in 5 of 9 rearrangement sites. No Wller sequencewas observed at these junction sites in all three testedtransformants.
123
240
Fig.4
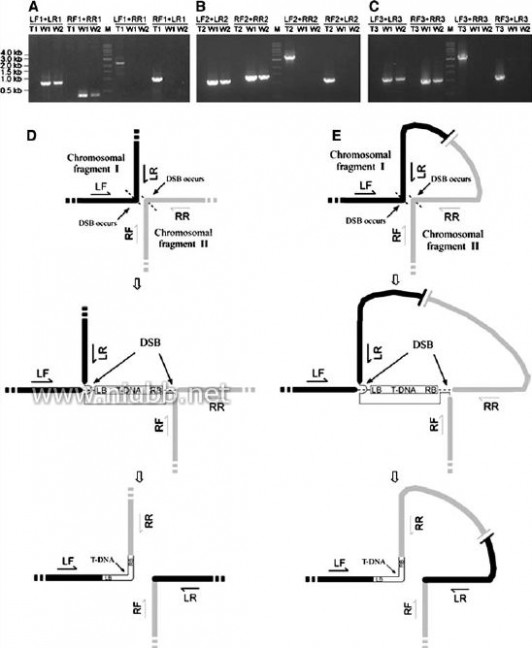
PCR analysis and pro-posed modes of chromosomal rearrangements. Each puta-tive transformant, 6001
(a), 4825 (b) or 438 (c) were analyzed by PCR using four sets of primer pairs (which relative positions were shown in d or e) as indicated on the top of each panel. Wild types Y34 and 70-15 were used as control. Original strains of templates were shown under primers. T1, T2, and T3 de-note transformant 6001, 4825, and 438, respectively; W1 and W2 denote Y34 and 70-15, respectively; M, 1kb DNA marker. Modes of chromo-somal rearrangement associ-ated with T-DNA integration are proposed based on molec-ular evidences of this study. When invading T-DNA mole-cule is physically close to host chromatins where two DSB events existed simultaneously, rearrangement might happen. d Translocation occurs (as transformant 438) if the two DSB events involved in rear-rangement derived from diVerent chromosomes. e Inversion occurs (as trans-formant 6001 and 4825) if the two involved DSB events originated from a same chromosome. DSB, double-stranded break
Curr Genet (2007) 51:233–243
Complex T-DNA integration patterns
Of the 623 RB Xanking sequences, 65 (10.4%) wereinvolved in long T-DNA integration. Southern blotanalysis (data not shown) with probes derived fromdiVerent regions of pBHt2 suggested that some trans-formants might contain at least one copy of the wholebinary vector. To test that speculation, Wve transfor-mants were selected for further assay of plasmidrescue. Genomic DNAs were digested with PstI andself-ligated. Plasmids were rescued from four transfor-mants but failed from one. Analysis of restrictionendonucleases digestions and PCR reactions indicatedthat these retrieved plasmids were the same as the orig-inal plasmid pBHt2 (data not shown), suggesting thatlong T-DNA with greater-than-unit-length of thebinary plasmid has been integrated into the genome ofM. oryzae.
Analysis of RB Xanking sequences also revealedthat 11.4% (71 of 623) of transformants containedtwo T-DNAs (one might be partial) co-integratedinto the genome of M. oryzae with head-to-tailfusions (direct repeat). At the junctions, rejoiningpatterns similar to ones described above for generalT-DNA integration (Fig.3, Table3) were alsoobserved. Most RBs (98.6%) showed full-lengthfusions, but one had 1bp deletion. Sometimes, LBsundergone deletions of 1–180bp, and full-length LBfusions were found in 44 (62.0%) transformants.Filler sequence (4.2%, with 1bp) and microhomology(2.8%, with 1–2bp) were also found but rarely at thejunctions.
123
Curr Genet (2007) 51:233–243Fig.5

Southern blot analysis of transformant 6001. I Schematicdiagram of the chromosomal inversion event in transformant6001. The arrow denotes the orientation of the DNA fragment in-volved in inversion. Left panel represents the chromosomal frag-ment before inversion. Predicted T-DNA insertion sites and therestriction site of ApaI (short vertical line) are indicated. Rightpanel represents the inversion of chromosomal segment in trans-
Discussion
In this study, we have analyzed the T-DNA insertionpatterns in the genome of M. oryzae comprehensively.Although T-DNA integration into M. oryzae follows asimilar mechanism as previously described in plantsand yeast, more precise and less complex integrationpatterns have been observed.
Single-copy T-DNA insertion events were observedmore frequently (72.6%) in M. oryzae than that in rice(36–49%) (Sallaud etal. 2003; Sha etal. 2004). Theaverage number of T-DNA insertion is 1.36 in M. ory-zae, compared with 1.5 in Arabidopsis (Alonso etal.2003) and 1.76–2.0 in rice (Sallaud etal. 2003; Sha etal.2004).
Transferred DNA integrates into M. oryzae with ahigher frequency of canonical RB than that into plants.Canonical RBs were observed in up to 92% of theinserted T-DNAs, compared with 19–55% in Arabid-opsis (Brunaud etal. 2002; Forsbach etal. 2003) andrice (Kim etal. 2003), which implies that in plants,more complicated modiWcations may occur at RB dur-ing the process from VirD2 departure to integration. Itshould be considered that, the frequencies of trunca-tion at borders were probably underestimated to somedegree, because Xanking sequences can not be recov-ered successfully by TAIL-PCR from those transfor-mants containing T-DNA borders truncated with morethan 100bp in size, which is the length between the ter-tiary speciWc primers (MR3 or ML3) and the canonicalT-DNA borders (RB or LB).
Transferred DNA integrations into plants mostlyinduce small fragment deletions at host genomic targetsites. Comparing with T-DNA lines of Arabidopsis,241
formant 6001. The shaded segments represent regions immediateoutside of the inversion fragment. II Genomic DNAs of transfor-mant 6001 and wild type strains Y34 and 70–15 were digested withApaI and hybridized with probes (a–f) illustrated in I. Each probe(a–f) used in Southern blot analysis was located on chromosomewith black bars
which have deletions typically spanning 11–100bp insize, smaller deletions with 1–20bp were frequentlyobserved in transformants of M. oryzae. The frequencyof exact joining (simple ligation without Wller sequenceand microhomology) between T-DNA border and hostgenomic DNA in ATMT of M. oryzae (RB junction,67.8%; LB junction, 25.8%) was higher than that inrice (Kim etal. 2003) and Arabidopsis (RB junction,8.5–39.6%; LB junction, 4.3–4.9%) (Forsbach etal.2003). Filler sequences were observed less frequentlyin transformants of M. oryzae, compared with those ofplants.
Long T-DNA transfer may result from either skip-ping the left T-DNA border or initiation of T-DNAtransfer from the LB to bring vector backbonesequences into plant cells (Kononov etal. 1997). LongT-DNA integrations into M. oryzae were found in10.4% of the cases. Higher frequencies (25.5–33.2 and33–62%, respectively) of that insertion pattern havebeen described previously in rice and Arabidopsis(Wenck etal. 1997; Yin and Wang 2000; Sha etal.2004). For M. oryzae, ATMT is performed by directco-culture of A. tumefaciens with conidia, while tissueculture is usually used for rice and Xower Wltration forArabidopsis. It is not clear whether distinct approachesresults in some diVerences of T-DNA integration pat-terns in these organisms.
Transferred DNA insertion in one site with tworepeat fusions was observed in plants. Before integra-tion, the two separate T-DNAs were supposed to ligateeach other (De Neve etal. 1997; De Buck etal. 1999).In M. oryzae transformants, co-integration of two T-DNAs in direct repeat conWguration was observed.Microhomology was found rarely at junctions between
123
242T-DNA ends, which implies that the fusions betweenRB and LB T-DNA ends seemed to result from asimple ligation of blunt double stranded ends mostly.Although T-DNA fusions in head-to-head (i.e., RB–RB)conWguration were found frequently in transformationof plants (De Neve etal. 1997; De Buck etal. 1999), noone of that insertion pattern was observed in ourobtained RB Xanking sequences. One of the reasonsmay be due to the application of TAIL-PCR to recoverXanking sequences. Those inverted repeats seem not tobe ampliWed by PCR. Otherwise, inverted repeats areunstable in many organisms, and therefore escapeselection (De Neve etal. 1997).
It was proposed that DSBs might have a signiWcantrole in T-DNA integration (TzWra etal. 2003). Thegenome of eukaryotic cells is under constant attack byagents from environment or within, and DSBs arecreated frequently (Hill 1999). There are two princi-pal mechanisms, homology-directed repair and non-homologous end-joining, by which cells can repairDSBs. Naturally occurring DSBs may act as “baits” toattract the incoming T-DNAs to the sites of their inte-gration. Before integration, the single-stranded T-DNAis converted into a double-stranded intermediatemostly (TzWra etal. 2003). The DNA-repair machineryof host cell recognizes double-stranded T-DNA mole-cules as genomic DSBs probably and mediates theirintegration by a DNA-repair mechanism.
Indeed, patterns associated with T-DNA integrations,such as occurrence of microhomologies or Wller sequencesat junction sites, small fragment deletions or duplica-tions of host junction sequences, even chromosomalrearrangements, are also found during the process ofDSB repair (Daley etal. 2005). Double stranded T-DNAmolecules may be recognized as genomic DSBs byhost DNA-repair machinery, which explains why twoT-DNAs can be fused in inverted- or direct-repeatmodes and co-integrate into host genome.
Based on our experimental data, we propose thatchromosomal rearrangements associated with T-DNAintegrations, including translocation and inversion, aremainly, if not all, mediated by DSB repair machinery(Fig.4d, e). When invading T-DNA molecule is steri-cally close to host chromatins where two DSB eventsexisted simultaneously, rearrangement may happen. Ifthe two DSB events involved in rearrangement derivedfrom diVerent chromosomes, translocation occurs(Fig.4d). Otherwise, inversion occurs (Fig.5e).
Oleinick etal. (1994) reported that decondensedchromatin was more susceptible to DSB-inducingagents, which explains why T-DNA is inclined to inte-grate into transcriptionally active regions where chro-matin decondensation occur more frequently.
123
Curr Genet (2007) 51:233–243
Transferred DNA integrates into the genome of M.oryzae with more precise and less complex patternsthan that into plants, which probably reXect the deli-cate diversity existing in DSB-repair machinery amonghosts, meanwhile facilitate genetic analysis of transfor-mants and identiWcation of genes disrupted by T-DNAintegration. T-DNA is inclined to integrate into trans-criptionally active regions, and EST sequences can befound in half of the insertion-site surroundingsequences with up- and down-stream 1kb in size,respectively (data not shown), which implies thatATMT can be used eVectively to study gene functionsby forward genetics. Together with advantagesdescribed previously (Rho etal. 2001), ATMT will bean eVective tool for functional genomics research ofnot only M. oryzae but also other Wlamentous fungi,since more and more species have been or will besequenced. More than 30 mutants with obvious pheno-types, such as pathogenicity-deWcient or abnormal con-idiation, have been identiWed from our T-DNA taggedpopulation. These genetic materials will be very usefulto elucidate biology and pathology of rice blast.
AcknowledgmentsThis project was supported by the Ministryof Science and Technology of China (Grant 2002BA711A15).
References
Alonso JM, Stepanova AN, Leisse TJ, Kim CJ, Chen H, Shinn P,
Stevenson DK, Zimmerman J, Barajas P, Cheuk R, Gadri-nab C, Heller C, Jeske A, Koesema E, Meyers CC, Parker H,Prednis L, Ansari Y, Choy N, Deen H, Geralt M, Hazari N,Hom E, Karnes M, Mulholland C, Ndubaku R, Schmidt I,Guzman P, Aguilar-Henonin L, Schmid M, Weigel D, CarterDE, Marchand T, Risseeuw E, Brogden D, Zeko A, CrosbyWL, Berry CC, Ecker JR (2003) Genome-wide insertionalmutagenesis of Arabidopsis thaliana. Science 301:653–657Brunaud V, Balzergue S, Dubreucq B, Aubourg S, Samson F,
Chauvin S, Bechtold N, Cruaud C, DeRose R, Pelletier G,Lepiniec L, Caboche M, Lecharny A (2002) T-DNA integra-tion into the Arabidopsis genome depends on sequences ofpre-insertion sites. Embo Rep 3:1152–1157
Castle LA, Errampalli D, Atherton TL, Franzmann LH, Yoon
ES, Meinke DW (1993) Genetic and molecular characteriza-tion of embryonic mutants identiWed following seed transfor-mation in Arabidopsis. Mol Gen Genet 241:504–514
Chen S, Jin W, Wang M, Zhang F, Zhou J, Jia Q, Wu Y, Liu F,
Wu P (2003) Distribution and characterization of over 1000T-DNA tags in rice genome. Plant J 36:105–113
Chilton MD, Que Q (2003) Targeted integration of T-DNA into
the tobacco genome at double-stranded breaks: new insightson the mechanism of T-DNA integration. Plant Physiol133:956–965
Daley JM, Palmbos PL, Wu D, Wilson TE (2005) Non-homolo-gous end joining in yeast. Annu Rev Genet 39:431–451
De Buck S, Jacobs A, Van Montagu M, Depicker A (1999) The
DNA sequences of T-DNA junctions suggest that complexT-DNA loci are formed by a recombination process resem-bling T-DNA integration. Plant J 20:295–304
Curr Genet (2007) 51:233–243
De Neve M, De Buck S, Jacobs A, Van Montagu M, Depicker A
(1997) T-DNA integration patterns in co-transformed plantcells suggest that T-DNA repeats originate from co-integra-tion of separate T-DNAs. Plant J 11:15–29
Dean RA, Talbot NJ, Ebbole DJ, Farman ML, Mitchell TK,
Orbach MJ, Thon M, Kulkarni R, Xu JR, Pan H, Read ND,Lee YH, Carbone I, Brown D, Oh YY, Donofrio N, JeongJS, Soanes DM, Djonovic S, Kolomiets E, Rehmeyer C, Li W,Harding M, Kim S, Lebrun MH, Bohnert H, Coughlan S,Butler J, Calvo S, Ma LJ, Nicol R, Purcell S, Nusbaum C,Galagan JE, Birren BW (2005) The genome sequence of therice blast fungus Magnaporthe grisea. Nature 434:980–986Forsbach A, Schubert D, Lechtenberg B, Gils M, Schmidt R
(2003) A comprehensive characterization of single-copy T-DNA insertions in the Arabidopsis thaliana genome. PlantMol Biol 52:161–176
Gheysen G, Villarroel R, Van Montagu M (1991) Illegitimate
recombination in plants: a model for T-DNA integration.Genes Dev 5:287–297
Hill MA (1999) Radiation damage to DNA: the importance of
track structure. Radiat Meas 31:15–23
Kim SR, Lee J, Jun SH, Park S, Kang HG, Kwon S, An G (2003)
Transgene structures in T-DNA-inserted rice plants. PlantMol Biol 52:761–773
K?hler F, Cardon G, P?hlman M, Gill R, Schieder O (1989)
Enhancement of transformation rates in higher plants bylow-dose irradiation: are DNA repair systems involved in theincorporation of exogenous DNA into the plant genome?Plant Mol Biol 12:189–199
Kononov ME, Bassuner B, Gelvin SB (1997) Integration of T-DNA binary vector ‘backbone’ sequences into the tobaccogenome: evidence for multiple complex patterns of integra-tion. Plant J 11:945–957
Lacroix B, TzWra T, Vainstein A, Citovsky V (2006) A case of
promiscuity: Agrobacterium’s endless hunt for new partners.Trends Genet 22:29–37
Liu YG, Whittier RF (1995) Thermal asymmetric interlaced
PCR: automatable ampliWcation and sequencing of insertend fragments from P1 and YAC clones for chromosomewalking. Genomics 25:674–681
Mayerhofer R, Koncz-Kalman Z, Nawrath C, Bakkeren G,
Crameri A, Angelis K, Redei GP, Schell J, Hohn B, Koncz C(1991) T-DNA integration: a mode of illegitimate recombi-nation in plants. Embo J 10:697–704
Michielse CB, Hooykaas PJ, van den Hondel CA, Ram AF
(2005) Agrobacterium-mediated transformation as a tool forfunctional genomics in fungi. Curr Genet 48:1–17
Mullins ED, Chen X, Romaine P, Raina R, Geiser DM, Kang S
(2001) Agrobacterium-mediated transformation of Fusariumoxysporum: an eYcient tool for insertional mutagenesis andgene transfer. Phytopathology 91:173–180
Nacry P, Camilleri C, Courtial B, Caboche M, Bouchez D (1998)
Major chromosomal rearrangements induced by T-DNAtransformation in Arabidopsis. Genetics 149:641–650
Ohba T, Yoshioka Y, Machida C, Machida Y (1995) DNA rear-rangement associated with the integration of T-DNA in to-bacco: an example for multiple duplications of DNA aroundthe integration target. Plant J 7:157–164
Oleinick NL, Balasubramaniam U, Xue L, Chiu S (1994) Nuclear
structure and the microdistribution of radiation damage inDNA. Int J Radiat Biol 66:523–529
243
Rho HS, Kang S, Lee YH (2001) Agrobacterium tumefaciens-mediated transformation of the plant pathogenic fungus,Magnaporthe grisea. Mol Cells 12:407–411
Sallaud C, Meynard D, van Boxtel J, Gay C, Bes M, Brizard JP,
Larmande P, Ortega D, Raynal M, Portefaix M, OuwerkerkPB, Rueb S, Delseny M, Guiderdoni E (2003) Highly eY-cient production and characterization of T-DNA plants forrice (Oryza sativa L.) functional genomics. Theor Appl Gen-et 106:1396–1408
Sambrook J, Fritsch E, Maniatis T (1989) Molecular cloning: a
laboratory manual, 2nd edn. Cold Spring Harbor Labora-tory, Cold Spring Harbor, New York
Sha Y, Li S, Pei Z, Luo L, Tian Y, He C (2004) Generation and
Xanking sequence analysis of a rice T-DNA tagged popula-tion. Theor Appl Genet 108:306–314
Soanes DM, Skinner W, Keon J, Hargreaves J, Talbot NJ (2002)
Genomics of phytopathogenic fungi and the development ofbioinformatic resources. Mol Plant Microbe Interact 15:421–427
Stachel SE, Timmerman B, Zambryski P (1986) Generation of
single-stranded T-DNA molecules during the initial stages ofT-DNA transfer from Agrobacterium tumefaciens to plantcells. Nature 322:706–712
Talbot NJ, Salch YP, Ma M, Hamer JE (1993) Karyotypic varia-tion within clonal lineages of the rice blast fungus, Magna-porthe grisea. Appl Environ Microbiol 59:585–593
Tinland B, Schoumacher F, Gloeckler V, Bravo-Angel AM,
Hohn B (1995) The Agrobacterium tumefaciens virulence D2protein is responsible for precise integration of T-DNA intothe plant genome. Embo J 14:3585–3595
TzWra T, Frankman LR, Vaidya M, Citovsky V (2003) Site-spe-ciWc integration of Agrobacterium tumefaciens T-DNA viadouble-stranded intermediates. Plant Physiol 133:1011–1023TzWra T, Li J, Lacroix B, Citovsky V (2004) Agrobacterium T-DNA integration: molecules and models. Trends Genet20:375–383
Valent B (1990) Rice blast as a model system for plant pathology.
Phytopathology 80:33–36
Virts EL, Gelvin SB (1985) Analysis of transfer of tumor-induc-ing plasmids from Agrobacterium tumefaciens to petuniaprotoplasts. J Bacteriol 162:1030–1038
Wang K, Stachel SE, Timmerman B, Montagu MV, Zambryski
PC (1987) Site-speciWc nick in the T-DNA border sequenceas a result of Agrobacterium vir gene expression. Science235:587–591
Wenck A, Czako M, Kanevski I, Marton L (1997) Frequent col-linear long transfer of DNA inclusive of the whole binaryvector during Agrobacterium-mediated transformation.Plant Mol Biol 34:913–922
Windels P, De Buck S, Van Bockstaele E, De Loose M, Depicker
A (2003) T-DNA integration in Arabidopsis chromosomes.Presence and origin of Wller DNA sequences. Plant Physiol133:2061–2068
Xu JR, Peng YL, Dickman MB, Sharon A (2006) The dawn of
fungal pathogen genomics. Annu Rev Phytopathol 44: 337–366
Yin Z, Wang G-L (2000) Evidence of multiple complex patterns
of T-DNA integration into the rice genome. Theor ApplGenet 100:461–470
Zeigler RS, Leong SA, Teng PS (1994) Rice Blast Disease. CAB
International, Wallingford
123
四 : The sign reads "In case of _____ fire, break the g
| The sign reads "In case of _____ fire, break the glass and push _____ red button." |
A. 不填; a B. 不填; the C. the; the D. a; a |
| B |
考点:
考点名称:定冠词定冠词的定义:
定冠词the 有this,that,these,those等意义,但较弱,用于单数或复数名词前,主要用来特指,使一个或几个事物区别于所有其他同名的事物。
定冠词通常位于名词或名词修饰语前,但放在both、all、double、half、twice等词之后。
如:All the students in the class went out.班里所有的学生都出去了。
定冠词的用法:
1、表示特指:
如:Look! A car has stopped there. The car is beautiful.瞧,有辆汽车在那儿停下了。那辆汽车可真漂亮。
Why not ask the teacher? 为什么不问问老师?
2、与单数可数名词连用表类别:
如:I hate the telephone. 我讨厌电话。
The cobra is dangerous. 眼镜蛇是危险的。
3、与某些形容词连用表示类别:
如:The rich are not always happier than the poor. 富人并不总是比穷人过得开心。
Theoldaremorelikelytocatchcoldthantheyoung.老年人比年轻人容易感冒。
4、用于独一无二的事物名词前:
如:The earth goes around the sun. 地球绕着太阳转。
The sky was blue and clear. 天空清澈湛蓝。
5、用于方向或方位等名词前:
如:He looked towards the east. 他朝东望。
Turn to the right at the second crossing. 在第二十字路口向右拐。
6、用于序数词或形容词的最高级前:
如:You will be the second to speak. 你第二个发言。
Autumn is the best season here. 秋季是这里最好的季节。
7、用于乐器名词前表示演奏:
如:He plays the piano very well. 他的钢琴弹得很好。
注:若不是从演奏角度来考虑,而是考虑乐器的实体,则不一定用定冠词:
He bought a piano for his son. 他为儿子买了部钢琴。
定冠词与不定冠词互换用法比较:
1、在形容词最高级前一般加定冠词。但有时却用不定冠词,这时它不表示“最”的意思,而表示“非常”“很”的意思。
如:This is the most important question of all. 这是所有问题中最重要的一个。
This is a most important question. 这时一个非常重要的问题。
2、在序数词前加定冠词,表示“第几”;加不定冠词则表示“又”“再”。
如:Will you be the firse to read the text? 你第一个读课文好吗?
Will you have a second try? 你再试一次好吗?
3、在有些短语中,用定冠词和不定冠词一样。
如:The number of our school students is about 1500. 我校学生人数约为1500人。
定冠词的用法口诀:
特指双熟悉,上文已提及;
世上独无二,序数最高级;
某些专有名,习语及乐器。
以上口诀归纳了用定冠词的一般情况,即:
①特指某些人或物
②谈话双方都熟悉的人或事
③上文已经提到的人或事
④世界上独一无二的事物前
⑤序数词回形容词最高级前
⑥某些专有名词前
⑦一些习惯短语(如:intheday等)中和乐器前(如:playtheviolin/piano)。
定冠词知识体系:
定冠词用法拓展:
1、用于姓氏的复数前,表示全家人或全家中两个或两个以上的人:
如:The Browns live next to us. 布朗一家就住在我们隔壁。
The Greens have no Children. 格林夫妇没有小孩。
2、用来代替前面已提到的人的身体部位或衣着等的一部分:
如:He hit me in the face. 他打我的脸。
He caught the thief by the collar. 他抓住小偷的衣领。
3、用于逢整十数词的复数名词前,指世纪中的年代或人的约略年岁:
如:He began to learn French in his fifties. 他五十多岁开始学习法语。
He went to Japan with his family in the sixties. 他在60年代带家人去了日本。
4、用于某些单数可数名词前,使意义抽象化,指其属性或功能等:
如:This colour is pleasant to the eye. 这颜色悦目。
He is fond of the bottle. 他喜欢喝酒。
5、表示计算单位,含有a, each, per 之类的意义:
如:He is paid by the hour (piece). 他拿计时(件)工资。
It sells at two dollars the pound. 这东西每磅卖两美元。
6、用于人名前,或特指、或比喻、或指其作品等;用于某些产品的名称前,指产品:
如:He likes the Picasso. 他喜欢毕加索的画。
Lu Xun has been known as the Gorky of China. 鲁迅人称中国的高尔基。
7、用于江、(运)河、海、洋以及山脉、群岛、半岛、海岛、海峡、沙漠等名称的前:
如:the Chang jiang River 长江
the Pacific(Ocean) 太平洋
the Suez(Canal) 苏伊士运河
①关于湖名前是否用冠词通常要分两种情况:
中国的湖名在英译时,其前通常加定冠词:
the West Lake 西湖,the Dong ting Lake洞庭湖。
而外国的湖名前,多数不加定冠词,少数加定冠词,视习惯而定:
Lake Success 成功湖,the Lake of Geneva日内瓦湖
②山名的构成有两种方式:
若用于“山名+Mountains”,其前常用定冠词:the Jing gang Mountains 井冈山;
若用于“Mount/Mt+山名”,则通常不用冠词:Mount Tai 泰山。
另外,若不出现mountain一词时,则通常要用冠词:theAlps阿尔卑斯山。
8、用于由普通名词或含有普通名词构成的专有名词 (如国名、地名、政党、团体、组织机构以及旅馆、商店、学校、医院、文娱场所、建筑物等)前:
如:the United Nations 联合国
the People's Republic of China 中华人民共和国
the National People's Congress 全国人民代表大会
注:大学名称的构成要注意以下情况:
①对于以地名命名的大学,通常有两种形式 (注意冠词的有无):
如:the University of London / London University 伦敦大学
②对于以人名命名的大学,通常只有一种表达(不用冠词):
如:Yale University 耶鲁大学
Brown University 布朗大学
零冠词的概念:
名词前没有定冠词、不定冠词、或任何限定词的现象。
零冠词的用法:
零冠词是指名词前面没有不定冠词、定冠词,也没有其他限定词的现象,零冠词的用法如下:
1、表示抽象概括意义时,不可数名词和复数名词使用零冠词:
例:Books are my best friends. 书是我的好朋友。
Water boils at 100℃. 水在摄氏100度沸腾。
比较:The water in this river is undrinkable. 这条河的水不可饮用。
2、专有名词通常使用零冠词:
例:Lu Xun is a great Chinese writer. 鲁迅是一位伟大的中国作家。
London is the capital of England. 伦敦是英国的首都。
China is a developing socialist country. 中国是一个发展中的社会主义国家。
3、按照习惯下列各类名词使用零冠词:
1)季节、月份、星期以及节假日等名词:
例:Summer begins in June in this part of the country. 这个地区夏天从六月份开始。
We have no classes on Sunday. 星期日我们不上课。
There are a lot of people shopping at Christmas. 在圣诞节有很多人购买东西。
2)三餐饭菜的名词:
例:have supper 吃晚饭
come to dinner 去吃饭
3)语言、运动、游戏等名词:
例:She speaks Chinese. 她说汉语。
He plays football. 他踢足球。
Let's have a game of chess. 咱俩下盘棋吧。
4)在某些意义有改变的名词前要使用零冠词:
例:He has gone to school. (tolearn) 他去上学了。
They are in church just now. (to worship) 现在他们在做礼拜。
同样,in hospital是“住院(治疗)”,in prison是“服刑”,等等。
注意:如果在这类名词前加冠词,则表示去那里干与之无关的事:
例:go to the school 可理解为去学校看望人,而不是“学习”。
4、在表示职位、头衔、身份等名词前:
例:Professor Wang 王教授
Doctor Tompson 汤普生医生
President Lincoln 林肯总统
Dean of the English Department 英语系主任
零冠词的特殊用法:
1、用于物质名词前。物质名词表示泛指或一般概念时,通常用零冠词:
如:Water boils at 100℃. 水在摄氏100度沸腾。
Blood is thicker than water. 水浓于水(即亲人总比外人亲)。
表示泛指或一般概念的物质名词前,即使有一描绘性修饰语,仍用零冠词:
如:Don't eat rotten food. 不要吃腐烂的食物。
注:(1)若特指,物质名词前可用定冠词:
如:Is the water in the well fit to drink? 这井里的水能喝吗?
(2)表示一种、一杯、一场、一阵、一份等这样的概念时,可用不定冠词:
如:This is a very good wine. 这是一种很好的酒。
A coffee, please. 请给我来杯咖啡。
It was very cold and a heavy snow was falling. 当时天气很冷,正在下大雪。
2、用于抽象名词前。抽象名词表示泛指或一般概念时,通常用零冠词:
如:Do you like music? 你喜欢音乐吗?
Failure is the mother of success. 失败是成功之母。
表示泛指或一般概念的抽象名词前,即使有一描绘性修饰语,仍用零冠词:
如:I like light music very much. 我非常喜欢轻音乐。
注:(1)若特指,抽象名词前可用定冠词:
如:I like the music of Mozart. 我喜欢莫扎特的曲子。
(2)若表示一种、一类、一方面、那种、这种等这之类的概念时,可用不定冠词:
如:He lives a happy life. 他过着幸福的生活。
Physics is a science. 物理是一门科学。
(3)表示动作的一次、一例、一番等时,可用不定冠词:
如:Let me have a look. 让我看一看。
(4)表示与抽象名词意义相关的具体的人或事,可用不定冠词:
如:The book is a delight to read. 这书读来很有趣。
3、用于专有名词前。在通常情况下,专有名词前用零冠词:
如:Smith lives in London. 史密斯住在伦敦。
注:若特指,专有名词前有时也可用定冠词:
如:The Smith you're looking for no longer lives here. 你找的那个史密斯不住这儿了。
4、用于复数名词前。复数名词表示类别时,通常用零冠词:
如:Teachers should be respected. 教师应该受到尊重。
泛指不定量的人或物,也用零冠词:
如:We are students of ClassFive. 我们是五班的学生。
注:若特指,复数名词前应用定冠词:
如:The teachers should attend the meeting 教师应参加会议。
5、用于单数可数名词前。单数可数名词前用零冠词,主要有以下情况:
(1)用于表示家庭成员或nurse, cook, teacher等名词前:
如:Mother is not at home.妈妈不在家。
Ask nurse to put the child to bed 叫保姆孩子抱到床上去睡觉。
Teacher was satisfied with our work. 老师对我们的工作很满意。
(2)用于动词turn(变成),go(变成)后作表语的名词通常用零冠词:
如:He was a teacher before he turned writer. 他在成为作家之前是教师。
He has gone socialist. 他成了社会主义者。
(3)在让步状语从句的倒装句式中,单数可数名词通常用零冠词:
如:Child as he is, he knows a lot. 他虽然是个孩子,但已经很懂事了。
Teacher though he is, he can't knowe verything. 他虽然是老师,但也不可能什么都懂。
(4)单数可数名词用作呼语,通常用零冠词:
如:How is she, doctor? 医生,她怎么样?
Can you drive me to the station, driver? 司机,请送我去车站,好吗?
(5)在某些独立结构中通常用零冠词:
如:The teacher came in, book in hand. 老师走进教室,手里拿着书。
He was sitting in the chair, pipe in mouth. 他坐在椅子里,嘴里叼着烟斗。
(6)在“kind[sort]of+名词”这一结构中,名词通常用零冠词:
如:This kind of book is very interesting. 这种书很有趣。
He is the sort of person I really dislike. 他这种人我真不喜欢。
注:注意以下两句在含义上的差别:
Whatkindofcarisit?这是什么牌子的车?
Whatkindofacarisit?这种车质量如何?
(7)当单数可数名词含义抽象化具有形容词意味时,通常用零冠词:
如:The man was more animal than man. 那个人与其说是人,不如说是畜生。
I was fool enough to accep this offer. 我接受他的提议真是太傻了。
Are you man enough for this dangerous job? 你有勇气敢做这项危险的工作吗?
零冠词用法口诀:
下列情况应免冠,代词限定名词前;
专有名词不可数,学科球类三餐饭;
复数名词表泛指,两节星期月份前;
颜色语种和国名,称呼习语及头衔。
以上口诀主要概括了一般应“免冠”的几种情况,即:
①名词前已有作定语用的this、that、some、any、my等限定词。
②专有名词和不可数名词前。
③表示学科的(如:maths、Chinese、physics)名词前。
④球类活动的名词前及三餐总称前。
⑤复数名词表示泛指(一类人或事)时。
⑥节日、季节、星期、月份前。
⑦表示颜色(如:It's red/yellow.)、语种(如:speak English/Japanese)和国家的非全称名词(如:We live in China. They come from America.)。
⑧在称呼或表示头衔的名词前。
⑨某些习惯短语中(如:inbed、go to school 等)。
零冠词知识体系:
| 零 冠词 | 名词前面没有定冠词、不定冠词、和其他限定词的现象。 | 1、在某些专有或者抽象物质表示类别前 |
| 2、在表示类别复数名词前 | ||
| 3、在季节、月份、星期、三餐前 | ||
| 4、称呼语或表示头衔,职务的词前 | ||
| 5、学科和球类运动的名称前 | ||
| 6、名词前有代词或所有格 | ||
| 7、在某些固定词组中: at night by bus |
零冠词用法拓展:
(1)节假日、星期、月份、季节等通常用零冠词:
如:We had a good time on Christmas Day. 我们在圣诞节过得很愉快。
Monday comes before Tuesday. 星期二在星期一之后。
He was born in September, 1988. 他出生在1988年9月。
注:①我国用Festival构成的传统节日通常用定冠词:
如:the Spring Festival春节
the Mid-autumn Festival [theMoonFestival]中秋节
②若表示特指或心目中的专指,星期、月份、季节等名词前可用定冠词:
如:He went abroad in the September of 1988. 他于1988年9月出国。
He came on the Sunday and went away on the Monday. 他星期日来,星期一就走了。
③表示“某一个”或受描绘性定语修饰表示“某种”这样的意义时,节日、星期、月份、季节等名词也可用不定冠词:
如:My birthday happened to be on a Saturday. 我的生日碰巧是星期六。
She came round to see me on a sunny Sunday. 她在一晴朗的星期日来看了我。
We had a nice Christmas. 我们过了一个愉快的圣诞节。
④当季节名词不强调时间而强调季节的内涵时,通常用 the:
如:Winter is coming. 冬天要来了。(单纯指冬天的时间)
The winter is coming. 冬天要来了。(暗示寒冷)
(2)某些表示自然界时间变化现象的名词,与某些介词(如at, after, before, till, until, towards, from等) 构成短语时,通常用零冠词:
如:at day-break 在天亮时
beforedawn 在天亮前
at dusk 在黄昏时
after sunset 在日落后
after sunrise 在日出前
until sundown 直到日落
towards dark 天快黑时
at midnight 在半夜
from dawn till dusk 从早到晚
当day, night, evening, morning, afternoon 等表示抽象的时间概念时,通常用零冠词:
如:Night fell. 天黑了。
Evening came on. 夜幕来临。
It was late afternoon before he reached home. 傍晚时候他才到家。
(3)球类、三餐、茶点等名词前,通常用零冠词:
如:We play basketball in the afternoon. 我们下午打篮球。
What do you have for breakfast? 你早餐吃什么?
They were at tea when I called. 我来访时他们正在喝茶(吃茶点)。
注:①球类名词若不是作为一项体育活动看待,而是作为一个实实在在的东西来看待,则可以用冠词:
如:The basketball is mine. 这个篮球是我的。
He bought a basketball. 他买了一个蓝球。
②三餐饭被特指可用定冠词,若受形容词修饰且非特指,可用不定冠词:
如:The supper she cooked was delicious. 她做的晚餐很可口。
We had a good lunch at Uncle's. 我们在叔叔家吃了顿丰盛的午餐。
(4)当名词后接有数词表示顺序时,名词前通常用零冠词:
如:Lesson10 is more interesting than Lesson11. 第10课比第11课更有趣。
There's a picture of a ship on page15. 在第15页有张一艘船的照片。
(5)公园、广场、学校、语言等名词前通常用零冠词:
如:Hyde Park 海德公园
Central Park(纽约) 中内公园
Zhong shan Park中山公园
Tian AnMen Square天安门广场
speak English 说英语
Beijing University 北京大学
注:当语言名词表特指意义或指某一语言中的对应词时,通常用定冠词:
如:the English spoken in America and Canada 在美国和加拿大讲的英语
What's the English for this? 这个东西用英语怎么说?
另外,在语言名词后加上language一词时,也要用冠词:the English language。
(6)表示学习、生活、娱乐等的单数名词,若表示相关的活动时,通常用零冠词:
如:go to school (bed, church, town, class, college, etc)去上学 (睡觉,做礼拜,进城,上课,上大学,等)
in bed (school, class, college, church, prison, hospital,etc) 在睡觉 (上学,上课,上大学,做礼拜,坐牢,住院,等)
be sent to hospital (prison) 被送往医院住院或治疗(关进监狱)
School is over at twelve. 12点放学。
注:①若不是指活动,而是指具体的实物,则要用冠词。比较:
如:go to the bed到床边去 (侧重指“床”这个实体)
go to bed 上床睡觉(侧重指与“床”有关的活动,即睡觉)
be in the school 在这所学校里 (侧重指“学校”这个地点)
be in school 在上学(侧重指与“学校”有关的活动,即读书)
②但是cinema, theatre是例外,它们表示相关活动时,其前要用定冠词:
如:He often goes to the cinema (theatre). 他经常去看电影(看戏)。
I prefer the cinema to the theatre. 我喜欢看电影,不喜欢看戏。
③有时定冠词和零冠词的选择与英美英语的不同习惯有关:
如:in hosptital (英) 住院
in the hospital (美) 住院
go to university (英)上大学
go to the university (美)上大学
at table (英)在吃饭
at the table(美)在吃饭
(7)某些用介词by构成的方式的短语通常用零冠词:
①表示乘坐交通工具:
如:by bus 乘公共汽车
by bike(bicycle) 骑自行车
by plane/byair乘飞机
by ship(boat) 坐船
by land 走陆路
by sea 从海路
②表示用通讯或通信等方式:
如:by phone 用电话
by telegram 用电报
by letter 用信件
by post 用邮寄
by radio 用无线电
by hand 用手工
(8)表示正式的或独一无二的头衔或职位等,在用作宾语、表语、补语或同位语时,通常用零冠词:
如:John is captain of the team. 约翰是足球队的队长。
He is head of the foreign languages department. 他是外语系主任。
注:尽管有时也有用定冠词的现象,但以零冠词为普通。
(9)单数可数名词紧密联系的平行结构,通常用零冠词:
如:They are brother and sister. 他们是兄妹。
Please pass me pencil and paper. 请把纸笔递给我。
Boy and girl came up to me together. 一个男孩和女孩一起向我走来。
(10)有些短语用零冠词和定冠词均可,只是含义不同:
如:out of question 毫无疑问
out of the question 不可能,不值得考虑的
keep house 料理家务
keep the house 呆在家里不外出
in charge of 负责,管理,主管
in the charge of 在…的管理(负责)之下
(11)许多习语用零冠词:
如:catch fire 着火
give way 让路
lose heart 灰心
move hosue 搬家
send word 捎信
take place 发生
by chance 偶然
catch sight of 看见
make use of 利用
61阅读| 精彩专题| 最新文章| 热门文章| 苏ICP备13036349号-1